The progress of our understanding and study of genomics is no different than other fields historically. In geography old maps would have you falling off the end of the world (cause you know it’s flat right?). Then as we became more informed about our world (and it helps that explorers didn’t fall off the end) our maps evolved and today we have google maps, google view, google directions…yes, be comforted in knowing….google is always there…watching….you.
When the first genomes were sequenced it was much the same thing…ok, we have a genome–now what. Some people though that discovering the genome and putting it together would decode the ‘language’ of life…not so much. In our process of understanding a genome, we conduct assembly, gene/ORF finding, assignation of motifs and regulatory areas if possible. Understanding genomes also requires theory just as differences in anatomy suggest adaptation in animals and similarity suggesting common origins; so is it with genomes. Similarity suggesting common origins and differences in sequence suggesting adaptation. Genomes provide a common ‘yardstick’.
Antonis Rokas
Vanderbilt University
Topic: Evolutionary Genomics
So, given we’ve been on high octane genomics using NGS for 10 days straight Antonis was merciful to us for this talk and after a brief introduction to evolutionary genomics gave us a good rundown of comparative functional genomics, populations genomics and phylogenomics.
He talks to us about looking at the genome and finding sense within some that something like this:
Into something like this:
So genomes provide a common yardstick for comparison work and with sequencing getting cheaper and more prolific and data is just streaming over us in waterfall fashion we can almost start diagnosing high throughput sequencing data pathologies…as well well know sequencing technology while fantastic is far from perfect and it takes an army of tools in order to ensure the data is clean and processed correctly to facilitate maximum resolution for interpretation.
So I’m going to approach this post in the form of vignettes:
Vignette 1: Domesticating fungus like we domesticate the dog…’HEEL FungIDO!’
Let’s get our fungal gears and head over to Aspergillus. Study of Aspergillus oryzae has led to interesting insights into domestication of my favorite sake fungus. Evolutionarily A. oryzae is closely related to his/her ugly cousin A. flavus–a nasty bugger that is an agricultural pest, aflatoxin producer and costs a lot in damage every year. What’s a aflatoxin? Well I know it’s a carcinogen and since it doesn’t sound like a spa treatment, I think I’ll stay away.
A. oryzae on the other hand assists in the production of sake, soy sauce (shoyu) and miso. It’s a non-aflatoxin producer and receives a big thumbs up from the USDA. It has the pathway for aflatoxin but it’s inactivated. Essentially you can think of A. oryzae as the domesticated version of A. flavus. In the study they analyzed 16 genomes, 8 oryzae and 8 flavus to discern how this process of domestication occurred. They used Illumina sequencing and obtained 12-30 million 80 bp reads that amounted to about 20x coverage across the genomes. They then untangled ~100,000 SNPs and to asked how many genetic populations they had and also did some gene cluster mapping in regions. This led to some RNA-seq work that elucidated how A. oryzae because atoxic. The reason was that in the sake making process yeast was put into the mix to break down sugar to alcohol and so A. oryzae has to become atoxic so as to not kill the yeast (S. cerevisia). In summary Antonis gives us the ‘road to domestication’.
Pretty cool and if you want to know the nitty gritty details of all this:
Vignette 2: ‘Tree of life’ delusions?
Richard Dawkins once was quoted saying that he believed there was a tree of life and that we’d discover all it’s branches by 2050. When I think of this in terms of bacteria my mind reels, we haven’t been able to scratch the surface of bacterial diversity or all the branch relationships within it. And as Antonis goes on to discuss it’s pretty difficult for fungi…yeast…just about anything in fact. Where do you draw the line or define ‘species’? At what point is your marker whether it be one gene or a whole genome accurate enough at reflecting the ‘true’ evolutionary history of your organism? What resolution genetically do you have to go to?
Give us some spells and voodoo! Sadly there are none.
Going back to a similar mantra repeated many times during the workshop, ‘it’s going to depend’. The problem lies in gene(s) selection. Fact is, many genes when you compare their phylogenetic trees are highly incongruent…for those of you who are tired, that means ‘they don’t match’. Antonis showed that incongruency is pervasive (35-48%) in mammals, insects…and worse in other organisms. In using yeast it was very difficult to resolve relationships among species with deeper branches. There is a lot of disagreement. Concatenation of genes helps but again, it’ll depend how many you concatenate and the selection of genes you choose to concatenate (adaptive? conserved? a mix of both?). He got a comment from a reviewer that seemed particularly appropriate in describing the conundrum we face in finding ‘the tree of life’.
“Plainly stated, taxonomists keep digging the same hole and falling down it; all that has changed over the years is the sophistication of the shovel…” ~Anon Reviewer
So what do we do with all this? Is phylogenetics worthless then? Absolutely not…but as with all these blog entries…lets throw some more considerations for your OCD tendencies:
- High bootstrap value does not mean ‘seal of approval’ for relationships in a tree. While it’s not a useless measure by any means, the more taxa you have in your tree the higher the bootstrap can go just inherently–it’s apart of the math involved in bootstrapping and the Rokas lab found this in their work.
- Be aware of deep branches. I’m not saying they aren’t ‘true’ but generally shorter branches with high support are easier to tease apart than longer branches. Try to get lots of taxa in (computational time permitting) to ‘fill in’ where you are getting longer branches to resolve the clades better.
- I (mel) generally run trees with lots of closely related species/taxa as much as possible, then zoom into my clades of interest. This helps in that I can have lots of taxa that help with resolution but I don’t have to print a tree with 100’s of taxa I ultimately don’t care about, I can zoom into the topology and focus on the relationships of interest.
- Gene trees can and will disagree many times because of duplication events, lineage sorting, HGT/recombination, gene loss etc. Try concatenation and I have a habit of avoiding genes near mobile elements like transposons because that increases the likelihood they’ve been transferred perhaps.
- Use other data like RNA-seq data or increasing genomic depth to clarify/confirm your sequences and increase depth to improve your confidence in the quality of the sequences underlying your tree.

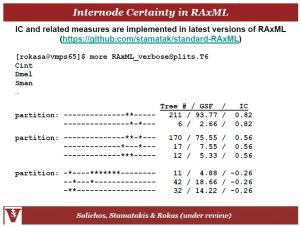
Antonis related to us his work in improving our confidence in tree nodes and therefore relationships between species in trees. They were able to develop a measure that gives you confidence in how many single genes trees match the concatenated tree based on sets of splits. So if you have a clade supported by a bootstrap value of 62, what about the other 38? Where do those topologies distribute in their confidence levels? So basically we are measuring the degree of conflict in internode data:
And this is implementable in RaxML:
So if you revisit the slide above the internode confidence (IC) of the 62 bootstrapped node is 0.59 meaning that the other splits were very low in terms of signal or confidence; so that node is actually well resolved. In contrast the node with bootstrap value of 52 only has an IC of 0.06 because there were so many splits that also had a decent amount of signal making evidence of that node quite weak.
Indeed, what can we do? This doesn’t ruin your ability to do phylogenetic analysis, rather it gives you more information in that now you know where the problem areas of your tree are and where you will have to do follow up work in order to resolve the relationships inferred by the tree or the alternate splits.
So in Summary:
- Few if any, of the gene trees are topologically identical to each other or to the phylogeny inferred by the concatenation.
- Concatenation analysis can be overconfident and misleading
- Internode support is inversely correlated with internode length and depth
- Selecting genes or gene tree partitions with strong signal reduces incongruence
“One can use the most sophisticated audio equipment to listen, for an eternity, to a recording of white noise and still not glean a useful scrap of information” ~Rodrigo et al. (1994) Chapter in: Sponge in Time and Space; Biology, Chemistry, Paleontology
For more information on this work check out:
- Salichos, L., and A. Rokas (2013). Inferring ancient divergences requires genes with strong phylogenetic signals. Nature 497: 327-331.
- Wired Magazine did an article about this work as well which is worth a read so check that out as well!
And if you want to learn more about the Rokas Lab and their fantastic work…Head over the Rokas Lab Website.
It was a pleasure having Antonis back!
Cheers,
…Dr. Mel