Before launching into today’s lecture, Konrad update us on the news that came out of Illumina. Illumina basically giving the finger to other technologies…ahem Ion Torrent…The Next-Seq 500 and HiSeq X10.
If you want to learn more about these newest developments along with additional thoughts on how they might compare to what is currently out there (and affordable) head over to Dr. Mick Watson‘s blog post on 7 reasons why getting a Next Seq 500 would be a strange choice. Nevertheless, Illumina is continuing it’s march toward the sea, furthing it’s domination in sequencing technology. William T. Sherman of the Union Army? Illumina?…Po-ta-to? Po-tah-toe?
Moving on…
Dr. Konrad Paszkiewicz
University of Exeter
Topic: Short Read Alignment
In general short read alignments are difficult because the shorter your read the less likely it is to match uniquely to a given reference or sequence of interest. Instead it’ll match to multiple places and you won’t be sure exactly where it goes.
Ok, so if it’s difficult to align short reads, why do people generate them? Well for one it’s cheaper. Additionally, for many applications a short read of about 50 bp is enough to work with; for example resequencing of small organisms, de novo analysis of bacterial genomes which are usually quite small compared to a human genome, ChIP-seq or digital gene expression.
Here is the current pipeline look, a little outdated but many of these programs are still utilized today and it gives you the general overview of what goes on depending on what you want to align.

Applications of short read alignment include genotyping that helps with methylation, SNPs, indels and to classify peaks required in experiments involving ChIP-seq or RNA-seq.
Given an alignment of a sequence that doesn’t match perfectly how do algorithms deal with this?
Algorithms will often times assign ‘penalties’ for mis-matches/substitutions or gaps in a given alignment. So a match would elicit a score of +1, a substitution would be -1 and a gap has a higher penalty at -2. These penalties can be changed in the programs.
ATCGATACG
ATCGA–ACG
The alignment above would then achieve a score of +6, because there are 8 exact matching giving you a score of +8 but you have 1 gap so you subtract two yielding a total alignment score of +6. The program’s algorithm builds matrices of these types of scores during alignment and at the end follows the path through the scores to construct the alignment with the best score.

There are two essential alignment strategies, global and local.
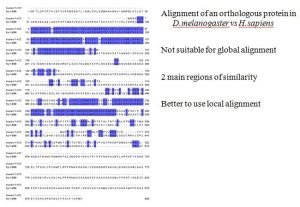
Global = When you are essentially attempting to align over the length of an entire length of two sequences which will most likely or hopefully be highly related with few mismatches or the algorithm will have difficulty.
Local = More tailored toward finding regions within a sequence that are highly similar and aligning around those then working out from there to attempt aligning the rest of the sequence which might be quite divergent.
Konrad went into amazing detail about how to make alignment matrices using Needleman-Wunsch (global alignment) and Smith-Waterman (local alignment). So I highly encourage you go back through his slides to review and appreciate how that is calcuated/done. Also for more details on Smith-Waterman aligning he linked up a great youtube video; admit it, you are on youtube already might as well check it out!
Both algorithms are ‘dynamic programming’ but are a bit impractical for ‘real world’ data which is quite a bit less clean or not as highly related perhaps and therefore these algorithms would take an incredibly long time to run. BLAST to the rescue…
BLAST
BLAST is an aligner that starts off with a ‘seed’ sequence which is ‘broken off’ from the sequence you are trying to match and once it finds a place on the reference where the match occurs at 100% identity it ‘extends’ out until the alignment score drops below 50%. Konrad illustrates this nicely in his slides.
Did you know BLAST has been around 22 years?! Since 1990 and it’s still one of the most widely used aligners.
But imagine using BLAST for NGS output. BLAST is great, but it takes 0.1-1.0 sec to spit out a result usually depending on the parameters and the sequence query you are trying to match. Ok, now toss 60 million illumina reads at it…I think it would give you the finger and shut down. It would take roughly 70 CPU days even on a multi-core system or using the upgraded implements to BLAST, it would last an ice age. So unfortunately, BLAST is not feasible for NGS data.
For NGS data to achieve alignment you have to make some concessions on sensitivity in an effort to get a dataset aligned before your hair turns grey.
So what are our options for NGS?
Well turns out there’s quite a few…I’m not going to link them all but all you have to do is toss the name of the program and ‘alignment’ or ‘aligner’ into Google and walah!
BLAT: older program that is still quite good and Dr. Kent wrote a great paper on it, highly recommended read.
SOAP
Bowtie2
MAQ
Shrimp2
CLCBio
So some of the adaptations that can be used to assist in alignment include allowing for mis-matches and gaps and/or using multiple seeds. Programs like Shrimp2 and CLCBio also allow you to reduce your ‘search space’ which can help speed up alignment. Here’s Konrad’s slide on the different aligners and what they allow/do not allow.
With absolute aligners (like BLAST) your seed has to be an ‘exact’ match…ever use BLAST and get the ‘no hits’ message that you wring your hands at whilst curing the computer? Well that occurs because none of your ‘seeds’ matched, perhaps you made them too long in size? Perhaps the organism you are trying to match is too divergent from what’s available to match it with? Try an aligner (like those mentioned) which allows for more flexibility (ie. allowing mis-matches and gaps). Aligners that allow you to adjust for mismatches are better for sequences where you are expecting 70% similarity (or greater). And as far as seed length…usually you should start with 20-30 bps but this can be adjusted depending on your read size.
So we’ve learned BLAST uses a seed approach and that it won’t work for NGS data because of the computational insanity that would ensue and we’ve listed some other options.
Programs like BWA, Bowtie and SOAP2 use a method called a suffix-prefix trie. It stores the ‘ends’ of the sequences and identical sequences. Not having to align sequences that are identical and would go to the exact same place saves time in alignment. This ‘compression’ of repeated bases is part of the Burrow-Wheeler transform if you want to get really geeky about it.
Considerations:
- Algorithms that use a hashed-seed approach take a lot of memory, about 30x slower, they are simpler in design and more sensitive.
- Algorithms that use a Suffix-Prefix Trie approach take much less memory, are about 30x faster, more complicated in design and are less sensitive.
- Also consider your read composition, if on average you are getting stretches of 3-4 bps of complete variability in comparison to what you are trying to match the read might be missed altogether. So even in ‘mis-match’ there can be a limit. That being said…I will make mention of GSNAP, an aligner where you can align very divergent sequences with no problem–it’ll allow any number of mismatches easily. It’s called a ‘SNP-tolerant’ alignment. So if you are banging your head against a brick wall with other programs take a look at GSNAP.
- Generally, and I know this is going to hurt for those large NGS datasets potentially, but if you have >2% divergence, it’ll be slower but you should probably use an aligner with a ‘hash’ approach. If you have <2% divergence then try to Trie-based ones.

from Konrad’s slides - To reduce homicidal aligning tendencies: quality clip your reads–don’t put junk in there, your alignment will take longer and cause more headaches. Also trim your reads for the same reason mentioned.
- Also pay attention to if certain aligners can do gapped alignments…for instance Bowtie cannot.
- Some aligners do not tolerate ambiguous bases and will score them as mismatches so be sure to remove all your N’s.
- A suggested paper to read with respect to all this is: David M et al., 2011. Shrimp2: sensitive yet practical short read mapping. Bioinformatics 27:1011. Granted it’s focused on Shrimp2 but figure 1 is quite nice in that it compares some of the latest aligners in terms of precision and recall and could serve as a great guide if you are unsure of where to start.

Other Considerations that can effect alignment:
- PCR duplicates: these can bias variant abundance. Never fear…SAMtools and Picard are here! They can identify PCR duplicates and remove them from the dataset. This is especially important for variant calling, so take note. Well constructed libraries should have < 2-3% duplicates.
- Base quality will impact your mapping/alignment: A good reference for this is Li and Homer’s paper in Bioinformatics.
- Methylation experiment alignment cannot be treated the same way as regular alignments due to the propensity of mismatches in un-methylated regions. So essentially you to have two references. Assuming your dataset is good, it will align/map to one of the references.
- What do you do with multiple mapping reads? Well, they could be real. They could be gene/chromosome duplication. Aligners will usually allow you to choose how to handle repeats. In the absence of ‘guidance’ they will usually assign them at random in the reference so you will get varying numbers of repeats in different areas usually just because of chance not because of some underlying biological mechanism.

You’ll notice novoalign actually has a better score, however this is not free nor open source, just to let you know.
- For RNA you will need a software package that can accoutn for splice variants. Try TopHat which is based on Bowtie or ERANGE. (http://woldlab.caltech.edu/rnaseq).
- If you have areas that were difficult to align because of indels, try realigning those areas specifically to see if you have improve your alignment.
- The Broad Institute also has the Genomic Analysis Toolkit so check that out as well.
Do refer to Konrad’s slides he has many great figures to depict all the considerations above that you need to take into account.
Always remember, no platform just spits out perfect data, it WILL have to be checked. You will have to check quality, possibly trim. Platforms like Ion Torrent and 454 Roche FLX have a tendency to shoot homopolymers. Illumina has issues with sequencing GC rich areas and will in general yield substitutions more than insertions.
Also your sequencing will only be as good as your library prep. If your library prep is biased your alignments will be biased.
OK…you have successfully mapped millions of reads, but what do you do with the unmapped reads? Well, first and foremost–they aren’t junk, so don’t toss them.
- They could be high quality, trimmed reads supposedly that just didn’t match your reference.
- They could also be errors (in which case toss ’em), contamination (possibly toss ’em if you don’t find the contamination interesting), reads of increased divergence, or transposons.
So what do you do with them…well you can
- Assemble them de novo and BLAST the contigs.
- Call ORFs and BLAST ORFs or you can use SwissProt a smaller curated database so it probably won’t take the lifetime that BLAST may promise. You could try BLAST2GO which will yield functional categorization. Or you could use PFam which is a collection protein families and can yield all kinds of awesome stuff.
You can also check out Nesoni which is a suite of tools focused on analyzing the alignment of reads to a reference genome. Well hopefully you have a nice start now on how to think about assembly, what might be right for your data and can begin the navigate the many options. Ultimately aside from picking an aligner that is appropriate for your data, user friendliness of the program will probably play a factor as well. Don’t get daunted by the programs, muscle through the documentation and if there is something you can’t get to work or don’t understand that’s what help pages, the literature, google, and emailing the creator are for!
So at the end of writing this post I asked Konrad how do I sum up Alignment algorithms in as few words as possible…Nick Loman immediately jumped in with the suggestion:
Heng Li Rocks.
Konrad agreed in Full…Heng Li Rocks…you’ll notice I cited several of his papers in this blog post…in case you missed them:
and in case you still doubt Heng Li’s enormity in the field of NGS computational analysis…
…oh and he created (along with Durbin) a somewhat AMAZING assembler called BWA.
Indeed…
Heng Li Rocks.




